DNA Methylation mediation of Adverse Outcomes in Type 2 Diabetes
1 Background
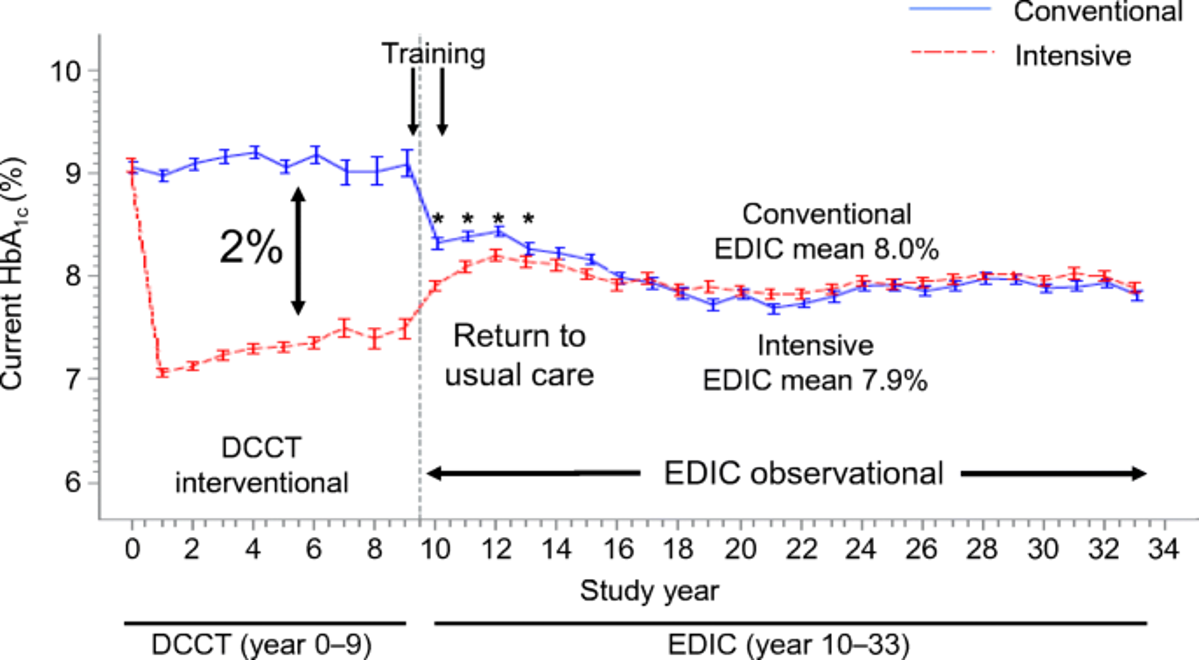
1.1 (Chen et al. 2016) DCCT-EDIC
- Examined DNA methylation data collected in the DCCT-EDIC study (\(n=1,441\)) using two sets of samples:
| Samples | Case | Control | Time of Sample Collection |
|---|---|---|---|
| Whole Blood DNA | 32 | 31 | EDIC Study baseline (1991-1993) |
| Monocytes | 31 | 30 | EDIC Study years 16-17 (2009-2010) |
where cases and controls were defined by:
Cases: DCCT conventional therapy group subjects with mean DCCT HbA1c > 9.1% and showing retinopathy or albuminuria progression by EDIC Study year 10.
Controls: DCCT intensive therapy group subjects with mean DCCT HbA1c < 7.3% and without complication progression by EDIC year 10.
Objective:
- identify the differentially methylated loci between the cases and controls
- whether or not the differential methylation levels persisted from baseline to year 16-17.
Finding:
- after adjustment, they identified 135 hypo-methylated CpG sites and 225 hyper-methylated CpG sites in cases vs controls.
- partly due to the differing cell-types in baseline and year 16-17 samples, they only identified 4 hypo-methylated and 8 hyper-methylated sites that were persistent.
1.2 (Chen et al. 2020) DCCT-EDIC
A larger cohort of 499 randomly selected participants: 125 each of the DCCT groups (Primary Intensive, Primary Conventional, Secondary Intensive, Secondary Conventional). One subject was excluded due to large differences in estimated white blood cell composition.
815,432 CpG sites, and identified 43 CpG sites associated with mean-DCCT HbA1c (how long and far back?) at a false discovery rate of \(<5\%\), which was further reduced to 11 CpG sites after Bonferroni adjustment (\(\alpha = 0.05\)).
Findings:
- “best combinations of multiple CpGs” selected from the top-ten HbA1c-associated CpG sites could explain the association between mean-DCCT HbA1c and the risk of developing diabetic complications, including
- \(71\%\) for proliferative diabetic retinopathy (PDR)
- \(73\%\) for severe nonproliferative diabetic retinopathy (SNPDR)
- \(68\%\) for clinically significant macular edema (CSME)
- \(97\%\) for albumin excretion rate \(>300\) per 24h (AER300),
- \(92\%\) for eGFR \(<60\) mL min–1 per 1.73 m2 (GFR60)
- \(84\%\) for eGFR slope.
- 4 HbA1c associated CpG sites as having methylation quantitative trait loci (meQTLs), from which one CpG site (cg08309687) had an identified causal effect on GFR60 development.
- “best combinations of multiple CpGs” selected from the top-ten HbA1c-associated CpG sites could explain the association between mean-DCCT HbA1c and the risk of developing diabetic complications, including
Pre-processing of methylation data was done using R package
minfiand association of historical HbA1c and CpG site methylation was estimated by multiple linear regression.
1.3 (Chen et al. 2024) Chen 2024
1.4 Joslin Kidney Study
1.5 Data
Subsample of 277 non-Hispanic white subjects with T1D AND had DKD at baseline
1.6 Others
A more recent work by (Kim et al. 2021) identified 8 differentially methylated CpG sites associated with type 2 diabetes in a case-control setting (232 cases, 197 controls), using subjects of East Asian ancestry.
Recent work (Yan et al. 2024) analyzed a cross-sectional cohort (\(n=399\)) seeking to identify differentially methylated sites among controls, and diabetes and hypertension attributed chronic kidney disease (CKD) patients, of which 136 were diabetic (unspecified type 1 or 2).
(Elliott et al. 2017; Juvinao-Quintero et al. 2023) sought to identify causal relationships between DNA methylation and type 2 diabetes ALSPAC-ARIES subjects (867 mothers and 385 fathers) with genetic and DNA methylation data to conduct analyses.
The same authors also authored a review (Juvinao-Quintero et al. 2019) of recent advances in causal effect estimation of DNA methylation on type 2 diabetes.
Glossary
DM: Diabetes Mellitus
EWAS: Epigenome-wide Association Study
DCCT: Diabetes Control and Complications Trial (1983-1993)
- EDIC: Epidemiology of Diabetes Interventions and Complications (1994-present), longitudinal monitoring of patients enrolled in DCCT
CpG site: cytosine/guanine
PDR: Proliferative Diabetic Retinopathy
SNPDR: Severe Nonproliferative Diabetic Retinopathy
CSME: Clinically Significant Macular Edema
AER: Albumin Excretion Rate
meQTL: Methylation Quantitative Trait Loci